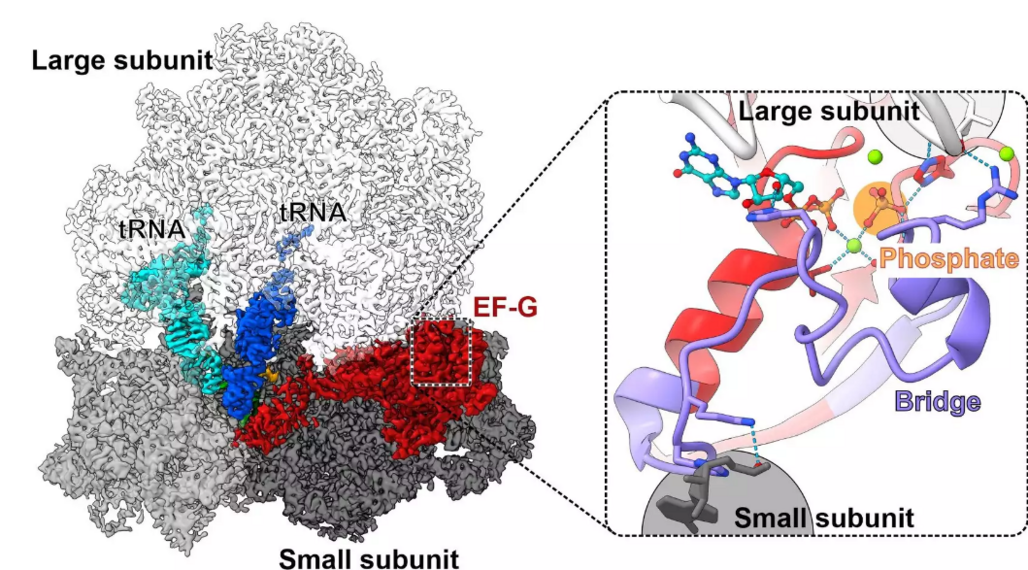
During protein synthesis, both transfer RNAs (tRNAs) and messenger RNA (mRNA) move rapidly through the ribosome. This movement is GTPase-powered by elongation factor G (EF-G). Niels Fischer, Marina Rodnina, and co-workers at the Max Planck Institute (MPI) for Biophysical Chemistry in Göttingen have visualized the steps of ongoing translocation by time-resolved cryo-electron microscopy (cryo-EM) revealing how a small local event drives large-scale movement in a huge RNA-protein complex. (Nature Communications, October 11, 2021)
The ribosome is a large macromolecular machine and often called ‘the protein factory of the cell’. It synthesizes proteins by translating the genetic information encoded in long strands of mRNAs. Each three nucleotides – the building blocks of the mRNA – form a so-called codon that encodes one amino acid. Amino acids, in turn, are the building blocks of proteins. The codons are decoded with the help of tRNA molecules. They bind specifically with one end to a codon on the mRNA and carry an amino acid on the other end. The ribosome has two main binding sites for tRNAs. When a tRNA delivers an amino acid, the ribosome catalyzes transfer of the amino acid from the tRNA at one binding site to the amino acid on the tRNA at the other binding site. In order to continue synthesis, the tRNAs and with them the mRNA have to be translocated by one codon through the ribosome. This molecular movement is powered by GTP hydrolysis by EF-G. However, how this works is a key unresolved question.
Time-resolved cryo-EM
Structural elucidation of EF-G-dependent translocation is challenging because it takes place within milliseconds. An interdisciplinary team of scientists around Niels Fischer and Marina Rodnina used biochemical methods to identify conditions under which translocation is slowed down and becomes accessible for time-resolved cryo-EM experiments. This allowed visualizing EF-G bound on the ribosome at the onset of translocation when GTP hydrolysis has occurred but the release of the resulting phosphate is delayed.
Phosphate release triggers a cascade of movements
Despite its large size, elements of the ribosome such as its large and small subunit and the bound tRNAs fluctuate rapidly between different states even in the absence of EF-G. The reported structures of the ribosome with EF-G before and after phospate release reveal key changes in the intricate networks of interactions that EF-G forms with the ribosome. Binding of EF-G-GTP stabilizes the rotated ribosome by bridging between the large and the small subunit. When phosphate is released, the GTP binding region in EF-G is remodeled, causing the interaction network to break down. This drives a large-scale rotation of EF-G and biases forward movement of ribosomal elements, which drag the tRNAs with them, eventually resulting in full translocation of the tRNAs and the mRNA.
These findings demonstrate at molecular detail how hydrolysis of a single GTP molecule orchestrates spontaneous fluctuations of a whole factory into force-generating molecular movement.
Contact
Prof. Marina Rodnina
Department of Physical Biochemistry
+49 551 201-2900
rodnina@...
Dr. Niels Fischer
Project Group Molecular Machines in Motion
+49 551 201-1306
nfische@...
Original publication
Petrychenko V, Peng BZ, de A P Schwarzer AC, Peske F, Rodnina MV, Fischer N
Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun (2021)
DOI